Caspases are a recently discovered group exists in the cytosol structurally related cysteine protease, which is an important common active site contains a cysteine. And it specifically breaks the peptide bond behind the acid residue of aspartic.
They are named caspase because of their specific cysteine protease activity, and caspase is derived from the abbreviation of cysteine aspartic acid specific protease. This name reflects the proteolytic characteristics of this family. And this high degree of specificity is rare in proteases. Due to this specificity, caspase can cleave certain proteins highly selectively. This cleavage only occurs at a few (usually only one) sites, mainly at the sites between the domains. The result of cleavage is to activate or inactivate a certain protein, but never completely degrade a protein.
The research on Caspase originated from the study of programmed cell death in C. elegans. During the development of the nematode, 131 cells will enter programmed death. The study found that there are 11 genes related to Programmed cell death (PCD), and the three key genes are ced-3, ced-4 and ced-9. Among them, ced-3 (ced, cell death defective) and Ced -4 is the apoptosis-determining gene of nematodes, called “killer gene”, if one of them is inactivated, all cells remain immortal throughout the development of nematodes; and ced -9 is a gene that inhibits apoptosis . ced -9 combines with ced -3 and ced -4 to form an “apoptotic body”, which controls the life and death of cells. When the cell does not receive the apoptosis signal, ced-9 combines with ced-4 to keep it in an inactive state, preventing downstream ced-3 precursor processing activation and cell apoptosis. Once the apoptotic signal is received, the apoptotic body is disintegrated, ced-4 oligomerizes, and ced-3 autocatalytic precursor is activated to play a role in promoting cell apoptosis. The study of programmed cell death in nematodes has promoted the study of apoptosis in other animals, especially mammals. Therefore, the homolog of Ced3 is found in mammalian cells. It is interleukin-1b converting enzyme (ICE), which catalyzes the activation of interleukin-1b, that is, cleaves IL-1b from its precursor. Overexpression of ICE and Ced3 in rat fibroblasts can cause apoptosis, indicating the similarity of ICE and Ced3 in structure and function. However, the phenotype of mice knocked out of the ICE gene was normal, and no significant changes in apoptosis were found. Further research revealed that another ICE member, later called apopain (CPP32 or Yama’s cysteine protease) catalyzes the cleavage of poly(ADP-ribose) Polymerase (PARP), resulting in cell apoptosis. Therefore, it is believed that apopain performs the same function as ced3 in nematodes. Apopain is known as the “death enzyme” and PARP is known as the “death substrate”. Apopain/CPP32/Yama were reported simultaneously by two laboratories in 1995, and the time difference was only two weeks. The mammalian homologue of Ced4 has not been discovered until 1997, it was proved to be apoptosis protease activating factor-1 (APAF-1). The mammalian homologue of Ced 9 was earlier proved to be BCL-2.
Caspase Type
It has been determined that there are at least 11 caspases:
Among these caspases, caspase 1 and caspase 11, and possibly caspase 4, are considered not to directly participate in the transduction of apoptosis signals. They are mainly involved in the activation of interleukin precursors. Caspase 2, caspase 8, caspase 9, and caspase 10 are involved in the initiation of cell apoptosis. Caspase 3, caspase 6, and caspase 7 are involved in the execution of apoptosis. Among them, caspase 3 and 7 have similar substrate and inhibitor specificities. They degrade PARP, DFF-45 (DNA fragmentation factor-45), which leads to inhibition of DNA repair and initiates DNA degradation. The substrates of caspase-6 are lamin A and keratin 18. Their degradation leads to the disintegration of the nuclear lamina and cytoskeleton.
Caspase Activation
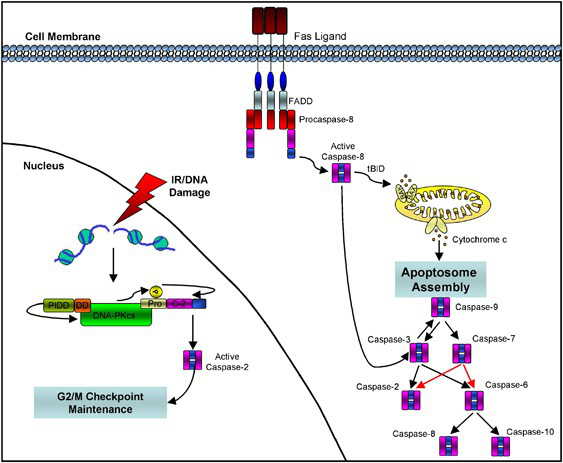
Due to the correlation between caspase and cell apoptosis, it exists in the cell in an inactive zymogen state, and can perform its function after activation. When the general protease is activated, only the N-terminal peptide is removed, while the activation of caspase requires cleavage at the aspartic acid site in the junction region of the two subunits, resulting in a two-subunit composition heterodimer, this is the enzyme with activity. Usually the N-terminal peptide is also removed during activation, but whether or not the N-terminal peptide is removed for caspase 7 has no effect on the activity. At present, it is believed that there is an upstream and downstream relationship between the initiator of apoptosis (caspase 2, 8, 9 and 10) and the performer (caspase 3, 6 and 7), that is, the initiator activates the performer. Apoptosis initiators (caspase 2, and 10) is activated by its multimeric form. Caspase 8 and 10 contain tandemly repeated “death effector” domains (DED), while caspase 2 and 9 contain different but similar caspase recruitment domains (CARD). These two domains can recruiting caspase 2, 8 And 10. In addition, caspase-8 terminates apoptosis, can cleave RIP1 and rip3, and effectively terminate necrosis.
