Post-transcriptional modifications of RNA molecules include methylation, acetylation deamination, isomerization, glycosylation, thiolation and pseudouracil modification among other types. Methylation represents the predominant post-transcriptional modification found within transfer RNA (tRNA) molecules. tRNA methylation has maintained its evolutionary conservation while also exhibiting dynamic and reversible properties within cellular environments. The stability of tRNA molecular conformation together with genome translation efficiency depend on this modification. Many diseases including neurological disorders and tumors result from defective tRNA methylation. The dynamic regulation of tRNA methylation is coordinated by three types of enzymes: The writer methyltransferase complex adds methyl groups while the eraser demethylase complex removes them and the reader RNA modification binding protein recognizes these modifications. This modification system plays a crucial role in cellular biological processes such as tumor metabolism and immune regulation while also shaping the tumor microenvironment through control of translation efficiency and mitochondrial function along with tRNA fragment generation. Through its impact on translation efficiency and mitochondrial function tRNA methylation modification also promotes tumor cell proliferation by controlling cell cycle-related protein expression. Cell cycle progression within cells is a critical biological function whose stoppage and progression rely entirely on cell cycle-related protein expressions. Tumor cells divide at a much faster rate than normal cells because they show significant dysregulation in cell cycle-related proteins. The altered activation states of cell cycle processes represent fundamental characteristics of tumors which scientists believe contribute majorly to tumor formation and progression. The potential to hinder tumor growth or boost drug effectiveness lies in the regulatory mechanisms of the cell cycle. The cell cycle process requires precise control through the synthesis and degradation of specific proteins including cell cycle checkpoint proteins together with DNA replication-related enzymes and cell division regulatory factors. Tumor cells exhibit abnormal cell cycle regulation that facilitates their rapid proliferation and substantial protein and energy requirements. Methylation modifications in tRNA impact tRNA expression levels while also controlling translation efficiency to fulfill tumor cells’ protein demands and affect mitochondrial function and energy supply. The modification of tRNA methylation is essential for controlling the cell cycle in tumor cells. The rapid advancement of high-throughput sequencing technology has enabled scientists to identify and report numerous tRNA methylation modifications and their modifying enzymes which control cell cycle regulation. Tumor prevention and treatment research anticipates tRNA methylation modifications and their associated enzymes to serve as new therapeutic targets.
tRNA Methylation Modification and Its Modifying Enzymes
tRNA functions as a crucial linker molecule during protein synthesis while it ensures precise genetic information transfer from parent cells to daughter cells. tRNA needs multiple post-transcriptional modifications to preserve its secondary “cloverleaf” structure while transforming into an adaptable “L-shaped” tertiary configuration through folding and splicing. TRNA can be categorized into cytoplasmic tRNA (ctRNA) and mitochondrial tRNA (mtRNA) based on their mature positions which results in distinct structural and functional differences between the two forms. The ctRNA molecule consists of approximately 73 to 93 nucleotides while maintaining a characteristic cloverleaf structure through its anticodon loop, DHU loop, TΨC loop, and variable loop which performs the primary role in cellular protein synthesis. mtRNA contains 57 nucleotides which enable it to carry out mitochondrial protein translation. Its abnormal modification affects mitochondrial function, thereby regulating energy metabolism.
tRNA methylation modification types mainly include m1A, N6-methyladenosine (m6A), N7-methylguanosine modification (m7G), N2-methylguanosine (m2G), N1-methylguanosine (m1G), N3-methylcytidine (m3C), N5-methylcytidine (m5C), 5-methyluridine (m5U), etc
Three Enzyme Types Controls tRNA Methylation Modification Level
tRNA methylation modification is catalyzed by specific modification enzymes: The methyltransferase complex known as the writer adds methyl groups to designated positions while the demethylase complex or eraser removes these methyl groups and the RNA modification binding protein or reader detects and binds to methylation sites which allows methylated RNA to fulfill specific functions. Writers, erasers, and readers work together to control tRNA methylation modification and affect essential biological mechanisms such as tumor metabolism and immune regulation through translation efficiency regulation and mitochondrial function management as well as tRNA fragment production. These enzymes play a critical role in both tumor development and advancement and show strong connections to negative prognoses.
Current research about tRNA methylation modification primarily investigates writers and erasers functions and mechanisms whereas reader studies remain sparse. Reader proteins such as the YTHDF family have been extensively studied and demonstrated to play a crucial role in cell cycle regulation through mRNA m6A modification. YTH domain family member 2 (YTHDF2) operates as a reader to regulate adipocyte cell cycle through its specific binding to m6A modifications. The removal of m6A modifications by fat mass and obesity-associated protein (FTO) from YTHDF2 binding sites affects the regulation of cyclin A2 and cyclin-dependent kinase 2 (CDK2) expression levels which in turn influences adipogenesis.
tRNA Methyltransferase Plays an Important Role in Cell Cycle Regulation
The m1A58 methyltransferase TRMT6-TRMT61A complex has been identified as a novel tumor-promoting factor that is upregulated in a variety of tumors, such as bladder cancer, glioma, gastrointestinal cancer, and hepatocellular carcinoma. It promotes cell proliferation and the transition of the G0/G1 phase of the cell cycle, and is associated with cell senescence. Studies have shown that TRMT61A deficiency significantly downregulates the m1A58 modification level of most tRNAs in T cells, reduces the translation rate of a variety of key cell cycle proteins, such as myelocytomatosis oncogene (MYC), and blocks the transition from G1 to S phase of the cell cycle. These results indicate that m1A modification catalyzed by the TRMT6-TRMT61A complex maintains the stability of tRNA structure, regulates translation efficiency, ensures the efficient synthesis of key cell cycle proteins, and promotes the smooth transition from G1 to S phase. Therapies that target hematopoietic stem cells’ self-renewal properties strike tumor stem cells with higher precision while safeguarding normal tissues from damage. The cell cycle quiescence status of HSCs helps them preserve their capacity for long-term self-renewal. The mammalian rapamycin complex 1 (mTOR complex 1, mTORC1) serves as a critical component for the metabolic processes and cell cycle advancement in hematopoietic stem cells. When the mTOR signaling pathway becomes abnormally activated HSCs enter the G1 phase of the cell cycle which leads to rapid proliferation loss of stemness and stability alongside a negative impact on the body’s health and potential acceleration of aging. Studies have shown that TRMT6 is closely related to the maintenance of HSC quiescence. HSCs lacking TRMT6 quickly leave the G0 phase and rapidly undergo apoptosis. TRMT6 deficiency causes the loss of function of Tsc1, a negative regulator of the mTOR signaling pathway. At this time, the mTOR signaling pathway is abnormally activated, promoting HSC proliferation and exhausting its self-renewal activity. These results show that TRMT6 has functional diversity in different cells, and its role and influence in cells vary depending on the cell type.
Post-transcriptional modifications of RNA molecules include methylation, acetylation deamination, isomerization, glycosylation, thiolation and pseudouracil modification among other types. Methylation represents the predominant post-transcriptional modification found within transfer RNA (tRNA) molecules. tRNA methylation has maintained its evolutionary conservation while also exhibiting dynamic and reversible properties within cellular environments. The stability of tRNA molecular conformation together with genome translation efficiency depend on this modification. Many diseases including neurological disorders and tumors result from defective tRNA methylation. The dynamic regulation of tRNA methylation is coordinated by three types of enzymes: The writer methyltransferase complex adds methyl groups while the eraser demethylase complex removes them and the reader RNA modification binding protein recognizes these modifications. This modification system plays a crucial role in cellular biological processes such as tumor metabolism and immune regulation while also shaping the tumor microenvironment through control of translation efficiency and mitochondrial function along with tRNA fragment generation. Through its impact on translation efficiency and mitochondrial function tRNA methylation modification also promotes tumor cell proliferation by controlling cell cycle-related protein expression. Cell cycle progression within cells is a critical biological function whose stoppage and progression rely entirely on cell cycle-related protein expressions. Tumor cells divide at a much faster rate than normal cells because they show significant dysregulation in cell cycle-related proteins. The altered activation states of cell cycle processes represent fundamental characteristics of tumors which scientists believe contribute majorly to tumor formation and progression. The potential to hinder tumor growth or boost drug effectiveness lies in the regulatory mechanisms of the cell cycle. The cell cycle process requires precise control through the synthesis and degradation of specific proteins including cell cycle checkpoint proteins together with DNA replication-related enzymes and cell division regulatory factors. Tumor cells exhibit abnormal cell cycle regulation that facilitates their rapid proliferation and substantial protein and energy requirements. Methylation modifications in tRNA impact tRNA expression levels while also controlling translation efficiency to fulfill tumor cells’ protein demands and affect mitochondrial function and energy supply. The modification of tRNA methylation is essential for controlling the cell cycle in tumor cells. The rapid advancement of high-throughput sequencing technology has enabled scientists to identify and report numerous tRNA methylation modifications and their modifying enzymes which control cell cycle regulation. Tumor prevention and treatment research anticipates tRNA methylation modifications and their associated enzymes to serve as new therapeutic targets.
tRNA Methylation Modification and Its Modifying Enzymes
tRNA functions as a crucial linker molecule during protein synthesis while it ensures precise genetic information transfer from parent cells to daughter cells. tRNA needs multiple post-transcriptional modifications to preserve its secondary “cloverleaf” structure while transforming into an adaptable “L-shaped” tertiary configuration through folding and splicing. TRNA can be categorized into cytoplasmic tRNA (ctRNA) and mitochondrial tRNA (mtRNA) based on their mature positions which results in distinct structural and functional differences between the two forms. The ctRNA molecule consists of approximately 73 to 93 nucleotides while maintaining a characteristic cloverleaf structure through its anticodon loop, DHU loop, TΨC loop, and variable loop which performs the primary role in cellular protein synthesis. mtRNA contains 57 nucleotides which enable it to carry out mitochondrial protein translation. Its abnormal modification affects mitochondrial function, thereby regulating energy metabolism.
tRNA methylation modification types mainly include m1A, N6-methyladenosine (m6A), N7-methylguanosine modification (m7G), N2-methylguanosine (m2G), N1-methylguanosine (m1G), N3-methylcytidine (m3C), N5-methylcytidine (m5C), 5-methyluridine (m5U), etc.
Three Enzyme Types Controls tRNA Methylation Modification Level
tRNA methylation modification is catalyzed by specific modification enzymes: The methyltransferase complex known as the writer adds methyl groups to designated positions while the demethylase complex or eraser removes these methyl groups and the RNA modification binding protein or reader detects and binds to methylation sites which allows methylated RNA to fulfill specific functions. Writers, erasers, and readers work together to control tRNA methylation modification and affect essential biological mechanisms such as tumor metabolism and immune regulation through translation efficiency regulation and mitochondrial function management as well as tRNA fragment production. These enzymes play a critical role in both tumor development and advancement and show strong connections to negative prognoses.
Current research about tRNA methylation modification primarily investigates writers and erasers functions and mechanisms whereas reader studies remain sparse. Reader proteins such as the YTHDF family have been extensively studied and demonstrated to play a crucial role in cell cycle regulation through mRNA m6A modification. YTH domain family member 2 (YTHDF2) operates as a reader to regulate adipocyte cell cycle through its specific binding to m6A modifications. The removal of m6A modifications by fat mass and obesity-associated protein (FTO) from YTHDF2 binding sites affects the regulation of cyclin A2 and cyclin-dependent kinase 2 (CDK2) expression levels which in turn influences adipogenesis.
tRNA Methyltransferase Plays an Important Role in Cell Cycle Regulation
The m1A58 methyltransferase TRMT6-TRMT61A complex has been identified as a novel tumor-promoting factor that is upregulated in a variety of tumors, such as bladder cancer, glioma, gastrointestinal cancer, and hepatocellular carcinoma. It promotes cell proliferation and the transition of the G0/G1 phase of the cell cycle, and is associated with cell senescence. Studies have shown that TRMT61A deficiency significantly downregulates the m1A58 modification level of most tRNAs in T cells, reduces the translation rate of a variety of key cell cycle proteins, such as myelocytomatosis oncogene (MYC), and blocks the transition from G1 to S phase of the cell cycle. These results indicate that m1A modification catalyzed by the TRMT6-TRMT61A complex maintains the stability of tRNA structure, regulates translation efficiency, ensures the efficient synthesis of key cell cycle proteins, and promotes the smooth transition from G1 to S phase. Therapies that target hematopoietic stem cells’ self-renewal properties strike tumor stem cells with higher precision while safeguarding normal tissues from damage. The cell cycle quiescence status of HSCs helps them preserve their capacity for long-term self-renewal. The mammalian rapamycin complex 1 (mTOR complex 1, mTORC1) serves as a critical component for the metabolic processes and cell cycle advancement in hematopoietic stem cells. When the mTOR signaling pathway becomes abnormally activated HSCs enter the G1 phase of the cell cycle which leads to rapid proliferation loss of stemness and stability alongside a negative impact on the body’s health and potential acceleration of aging. Studies have shown that TRMT6 is closely related to the maintenance of HSC quiescence. HSCs lacking TRMT6 quickly leave the G0 phase and rapidly undergo apoptosis. TRMT6 deficiency causes the loss of function of Tsc1, a negative regulator of the mTOR signaling pathway. At this time, the mTOR signaling pathway is abnormally activated, promoting HSC proliferation and exhausting its self-renewal activity. These results show that TRMT6 has functional diversity in different cells, and its role and influence in cells vary depending on the cell type.
Other recent studies have shown that no significant differences in tRNA m1A modification levels were found between young and old hematopoietic stem cells and their progenitors. However, inconsistent with the above findings, abnormally high expression of the TRMT6-TRMT61A complex upregulated tRNA m1A58 modification, which also led to HSC exhaustion and aging. The report proposed a hypothesis that the TRMT6-TRMT61A complex may not be a regulatory factor, but a type of signal marker. Its abnormal expression leads to abnormal mTOR signaling pathways and affects the functional state of HSCs. These results indicate that the role of tRNA m1A modification in the mechanism of HSC aging has not been fully elucidated, and its methyltransferase TRMT6-TRMT61A complex may have diverse functions in cells.
tRNA Demethylase Plays an Important Role in Cell Cycle Regulation
The synergistic effect of methyltransferase and demethylase is the key mechanism of dynamic and reversible RNA methylation modification. Methyl groups are added to RNA molecules under the action of methyltransferase and removed by demethylase. Therefore, the two are vividly called “writer” and “eraser”, both of which have substrate specificity. Compared with writers, erasers can bind to a wider range of substrates, covering mRNA, tRNA, rRNA and other RNA molecules. For example, ALKBH3 can act as an RNA demethylase and repair alkylation damage on DNA. The tRNA modification-related demethylases that have been identified so far all belong to the alkylated DNA repair protein B (Alkb) family, such as FTO and ALKBH 1/3/5/7. Most of these enzymes can act on multiple tRNA modifications, and each tRNA modification may be removed by multiple demethylases. tRNA demethylases play a potentially important role in the regulation of the cell cycle. Although many studies have shown that members of the demethylase family are involved in cell cycle regulation, the molecular mechanism by which they exert their effects through the tRNA modification pathway has not been fully elucidated.
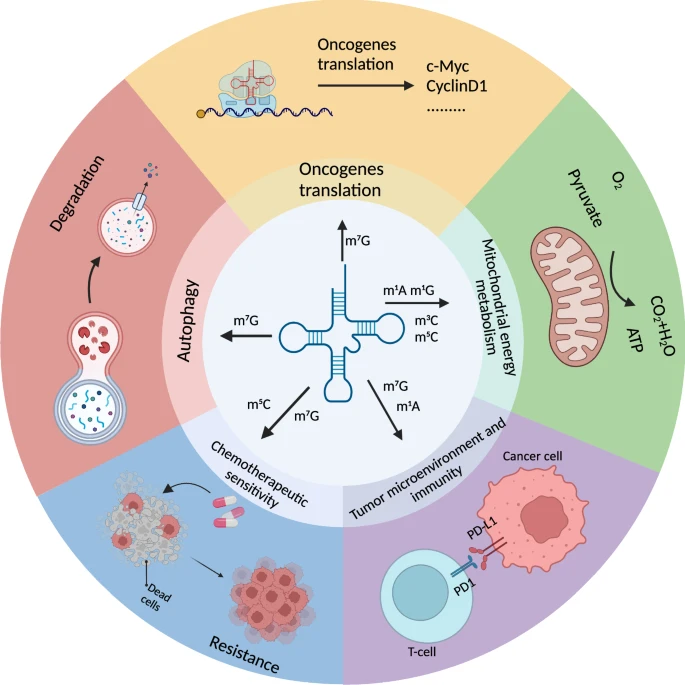
Figure 1. The biological function of tRNA methylation in cancer cells.
Other recent studies have shown that no significant differences in tRNA m1A modification levels were found between young and old hematopoietic stem cells and their progenitors. However, inconsistent with the above findings, abnormally high expression of the TRMT6-TRMT61A complex upregulated tRNA m1A58 modification, which also led to HSC exhaustion and aging. The report proposed a hypothesis that the TRMT6-TRMT61A complex may not be a regulatory factor, but a type of signal marker. Its abnormal expression leads to abnormal mTOR signaling pathways and affects the functional state of HSCs. These results indicate that the role of tRNA m1A modification in the mechanism of HSC aging has not been fully elucidated, and its methyltransferase TRMT6-TRMT61A complex may have diverse functions in cells.
tRNA Demethylase Plays an Important Role in Cell Cycle Regulation
The synergistic effect of methyltransferase and demethylase is the key mechanism of dynamic and reversible RNA methylation modification. Methyl groups are added to RNA molecules under the action of methyltransferase and removed by demethylase. Therefore, the two are vividly called “writer” and “eraser”, both of which have substrate specificity. Compared with writers, erasers can bind to a wider range of substrates, covering mRNA, tRNA, rRNA and other RNA molecules. For example, ALKBH3 can act as an RNA demethylase and repair alkylation damage on DNA. The tRNA modification-related demethylases that have been identified so far all belong to the alkylated DNA repair protein B (Alkb) family, such as FTO and ALKBH 1/3/5/7. Most of these enzymes can act on multiple tRNA modifications, and each tRNA modification may be removed by multiple demethylases.
tRNA demethylases play a potentially important role in the regulation of the cell cycle. Although many studies have shown that members of the demethylase family are involved in cell cycle regulation, the molecular mechanism by which they exert their effects through the tRNA modification pathway has not been fully elucidated.
| Cat. No. | Product Name | Application | |
| DMABB-JX274 | Rabbit Anti-5-methylcytosine (5-mC) monoclonal antibody, clone SN342 | ELISA, Dot, ICC, FC | Inquiry |
| DMABT-H21873 | Mouse Anti-5-Methylcytosine monoclonal antibody, clone 44E4 | Dot, IF, SB, ICC, IHC-P, FC | Inquiry |
| CABT-B354 | Sheep Anti-5-Methylcytosine polyclonal antibody | Dot, FC, ICC/IF, IHC, IP, WB | Inquiry |
| Cat. No. | Product Name | Application | |
| DMABB-JX352 | Rabbit Anti-2′-O-methylcytidine/Cm monoclonal antibody, clone BSD61740 | ELISA, Dot | Inquiry |
| DPABB-JX167 | Rabbit Anti-2′-O-methylcytidine/Cm polyclonal antibody | ELISA, Dot | Inquiry |
| Cat. No. | Product Name | Application | |
| DMABB-JX344 | Rabbit Anti-5-Carboxylcytosine (5-caC) Monoclonal Antibody, clone SN572 | Dot, ELISA | Inquiry |
| DPABB-JX157 | Rabbit Anti-5-Carboxylcytosine (5-caC) polyclonal antibody | Dot, IF/ICC | Inquiry |
| Cat. No. | Product Name | Application | |
| DPABB-JX159 | Rabbit Anti-3-Methylcytosine (3-mC) polyclonal antibody | Dot | Inquiry |